Self Supervised Learning of Electron Microscopy Images
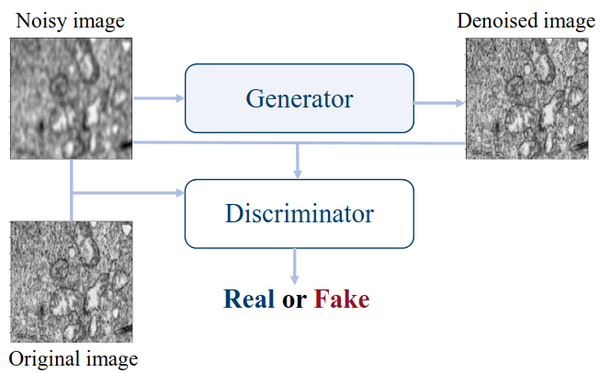
This research explores self-supervised learning using Generative Adversarial Networks (GANs) to enhance the analysis of electron microscopy (EM) images. Instead of relying on manually labeled data, the method leverages vast collections of unlabeled EM images to pretrain neural networks that learn rich, transferable representations of microscopic structures. Using the Pix2Pix conditional GAN architecture (Isola et al., 2017), we train a generator–discriminator pair to reconstruct clean, realistic EM images from noisy inputs, capturing underlying structural patterns and textures characteristic of nanoscale materials.
After pretraining on large unlabeled datasets such as CEM500K (Conrad et al., 2021), the pretrained models are fine-tuned on task-specific and domain-specific datasets like HRTEM Au nanoparticles and TEMImageNet. This process substantially improves performance and accelerates convergence across various downstream tasks—including semantic segmentation, denoising, noise and background removal, and super-resolution. Notably, the pretrained models achieve better accuracy and stability even with simpler architectures (e.g., U-Net, HRNet) compared to larger models trained from scratch.
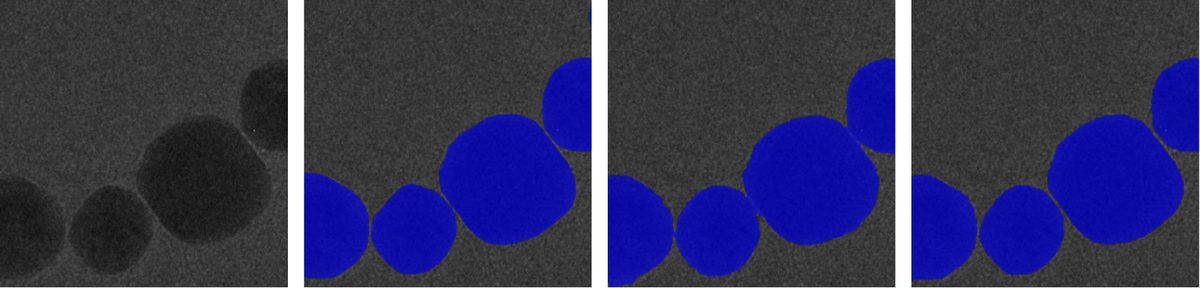
Our results show that self-supervised GAN-based pretraining can significantly reduce the dependency on annotated data and complex network architectures. It provides a framework for automated EM image interpretation, enabling more robust, data-efficient, and generalizable analysis pipelines for diverse imaging conditions and materials. This research establishes self-supervised pretraining as a powerful foundation for next-generation deep learning workflows in microscopy and materials characterization.
Related Publications:
- B. Kazimi, K. Ruzaeva and S. Sandfeld, "Self-Supervised Learning with Generative Adversarial Networks for Electron Microscopy," 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Seattle, WA, USA, 2024, pp. 71-81, doi: 10.1109/CVPRW63382.2024.00012.
Contact:
Dr.-Ing. Bashir Kazimi
Tel.: +49 241/927803-38
E-mail: b.kazimi@fz-juelich.de
