Tools and Reproducibility
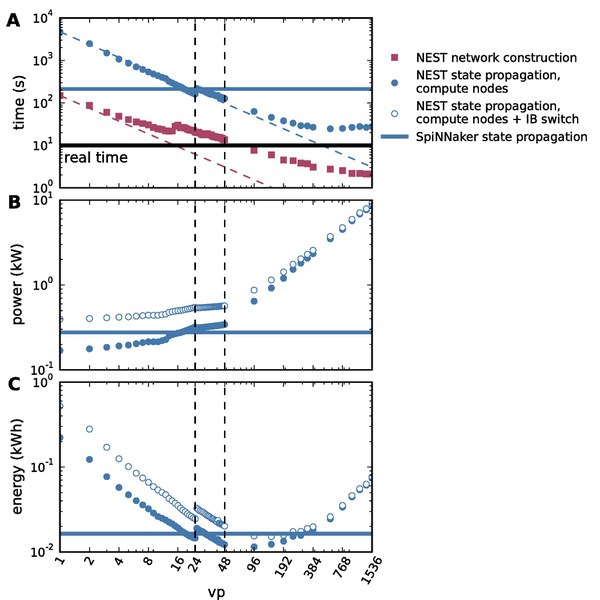
Copyright: Performance comparison of NEST and SpiNNaker for a cortical microcircuit model (van Albada et al., Front Neurosci 2018)
To enable computational neuroscientists to build on each other’s work in an efficient manner, making well-documented model and analysis code freely available is indispensable. Furthermore, obtaining neuroscientific findings relies on the development of appropriate data collation, simulation, analysis, and visualization tools. To support these efforts, we contribute to the development and publication of a variety of tools and model codes in computational neuroscience. In addition, we have proposed unified descriptions and graphical notations for commonly used connectivity concepts in neuronal network modeling to aid reproducibility.
Publications
- Senk J., Kriener B., Djurfeldt M., Voges N., Jiang HJ., Schüttler L., Gramelsberger G., Diesmann M., Plesser HE., van Albada SJ. (2022) Connectivity concepts in neuronal network modeling. PLoS Computational Biology 18(9):e1010086.
DOI: 10.1371/journal.pcbi.1010086 - Gleeson P., Cantarelli M., Marin B., Quintana A., Earnshaw M., Sadeh S., Piasini E., Birgiolas J., Cannon RC., Cayco-Gajic NA., Crook S., Davison AP., Dura-Bernal S., Ecker A., Hines ML., Idili G., Lanore F., Larson SD., Lytton WW., Majumdara A., McDougal RA., Sivagnanam S., Solinas S., Stanislovas R., van Albada SJ., van Geit W., Silver RA. (2019) Open Source Brain: A Collaborative Resource for Visualizing, Analyzing, Simulating, and Developing Standardized Models of Neurons and Circuits. Neuron 103: 395-411.e5.
DOI: 10.1016/j.neuron.2019.05.019 - van Albada SJ., Rowley AG., Senk J., Hopkins M., Schmidt M., Stokes AB., Lester DR., Diesmann M., Furber SB. (2018) Performance Comparison of the Digital Neuromorphic Hardware SpiNNaker and the Neural Network Simulation Software NEST for a Full-Scale Cortical Microcircuit Model. Frontiers in Neuroscience 12:291.
DOI: 10.3389/fnins.2018.00291 - Hagen E., Dahmen D., Stavrinou ML., Lindén H., Tetzlaff T., van Albada SJ., Grün S., Diesmann M., Einevoll GT. (2016) Hybrid Scheme for Modeling Local Field Potentials from Point-Neuron Networks. Cerebral Cortex 26:4461–4496.
DOI: 10.1093/cercor/bhw237. - Maksimov A, van Albada SJ. , Diesmann M. (2016) [Re] Cellular and Network Mechanisms of Slow Oscillatory Activity (<1 Hz) and Wave Propagations in a Cortical Network Model. Rescience.
DOI:10.5281/zenodo.161526 - Bakker R., Tiesinga P., Kötter R. (2015) The Scalable Brain Atlas: Instant Web-Based Access to Public Brain Atlases and Related Content. Neuroinformatics 13:353–366.
DOI: 10.1007/s12021-014-9258-x. - Nowke C., Hentschel B., Kuhlen T., Schmidt M., van Albada SJ., Eppler JM., Bakker R., Diesmann M. (2013) VisNEST – interactive analysis of neural activity data. IEEE BioVis 65-72.
DOI: 10.1109/BioVis.2013.6664348 - Bakker R., Wachtler T., Diesmann M. (2012) CoCoMac 2.0 and the future of tract-tracing databases. Frontiers in Neuroinformatics 6:30.
DOI: 10.3389/fninf.2012.00030. - Crook SM., Bednar JA., Berger S., Cannon R., Davison AP., Djurfeldt M., Eppler J., Kriener B., Furber S., Graham B., Plesser HE., Schwabe L., Smith L., Steuber V., van Albada SJ. (2012) Creating, documenting and sharing network models. Network: Computation in Neural Systems 23:131–149.
DOI: 10.3109/0954898X.2012.722743. - van Albada SJ, Robinson PA (2007) Transformation of arbitrary distributions to the normal distribution with application to EEG test-retest reliability. J Neurosci Meth 161: 205-211
DOI: 10.1016/j.jneumeth.2006.11.004
Book chapters
- van Albada SJ., Pronold J., van Meegen A., Diesmann M. (2021) Usage and Scaling of an Open-Source Spiking Multi-Area Model of Monkey Cortex. In: Amunts K., Grandinetti L., Lippert T., Petkov N. eds. Brain-Inspired Computing. Cham : Springer, Lecture Notes in Computer Science 12339, 47-59.
DOI: 10.1007/978-3-030-82427-3_4 - Senk J., Yegenoglu A., Amblet O., Brukau Y., Davison A., Lester DR., Lührs A., Quaglio P., Rostami V., Rowley A., Schuller B., Stokes AB., van Albada SJ., Zielasko D., Diesmann M., Weyers B., Denker M., Grün S. (2017) A Collaborative Simulation-Analysis Workflow for Computational Neuroscience Using HPC. In: Di Napoli E, Hermanns M-A, Iliev H, Lintermann A, Peyser A eds. High-Performance Scientific Computing. Cham: Springer International Publishing, 243–256.
DOI: 10.1007/978-3-319-53862-4_21.
Software
CoCoMac: Collation of Connectivity data on the Macaque brain
Scalable Brain Atlas: Web-based interactive brain atlases for macaque, mouse, rat, human, and more
NeuronsReunited neuron database viewer: Viewer for locations and morphologies of long-range projection neurons in mouse'
Potjans & Diesmann (2014) cortical microcircuit model on Open Source Brain
NEST code by Maksimov et al. (2016) reproducing results from Compte et al. (2003) Cellular and Network Mechanisms of Slow Oscillatory Activity (<1 Hz) and Wave Propagations in a Cortical Network Model
Collaborators
Prof. Paul Tiesinga, Radoud University Nijmegen, the Netherlands
Prof. Gaute Einevoll, Norwegian University of Life Sciences & University of Oslo, Norway
Prof. Hans Ekkehard Plesser, Norwegian University of Life Sciences, Norway
Dr. Stefan Mihalas, Allen Institute, USA
Funding
BMBF (Award ID 2424124) as part of the NSF/NIH/DOE/ANR/BMBF/BSF/NICT/AEI Collaborative Research in Computational Neuroscience Program (2025-2030)
Horizon Europe Programme under the Specific Grant Agreement No. 101147319 (EBRAINS 2.0 Project) (2024-2026)
European Union Horizon 2020 research and innovation program (Grant 945539, Human Brain Project, SGA3) (2020‒2023)
European Union Horizon 2020 research and innovation program (Grant 785907, Human Brain Project, SGA2) (2017‒2020)
