New model brings astrocytes into large-scale brain simulations
A new study that started in the Human Brain Project (HBP) - and was enabled by EBRAINS - introduces a framework for including astrocytes into large-scale simulations of brain networks
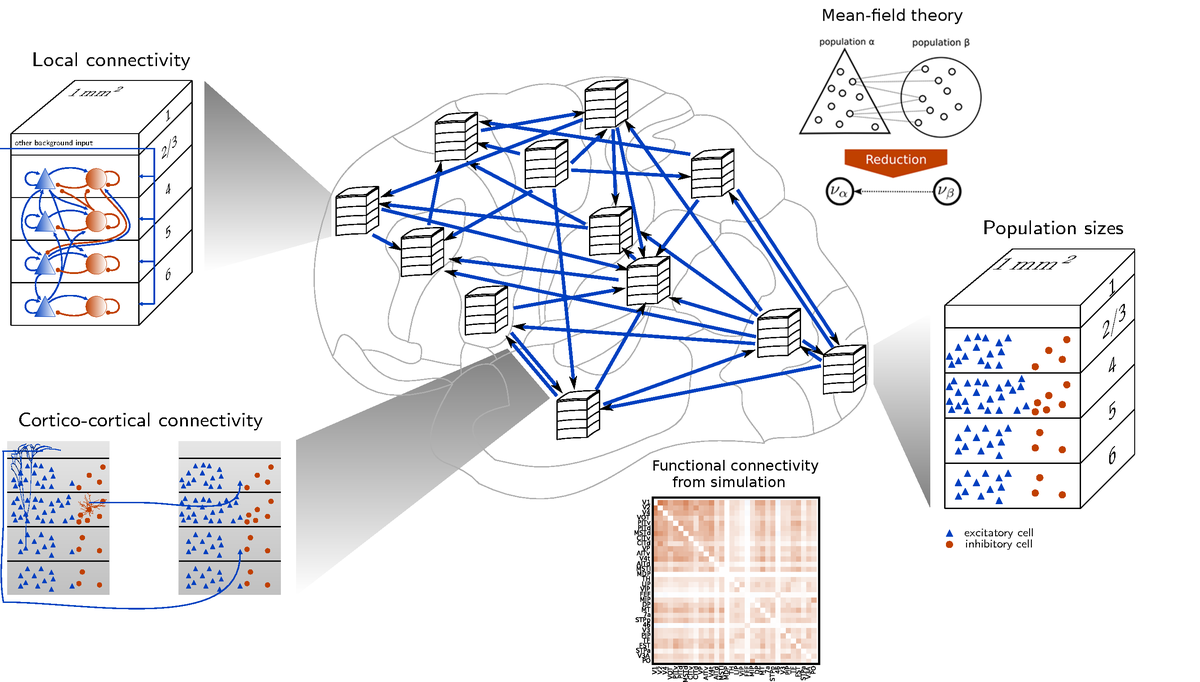
The work group led by Prof. Dr. Sacha van Albada studies the architecture and connectivity of brain circuits as the basis for neural network models at the resolution of neurons and synapses that relate structure to dynamics.
To build network models following the construction principles of the brain, the group combines the compilation of experimental anatomical data, for instance using the CoCoMac database, with statistical data prediction strategies. These strategies, including in particular predictive connectomics, yield new insights into the principles governing brain structure. The main focus of the research is on the cerebral cortex of mammals, characterizing its multi-scale structure using concepts like areas, layers, and neural populations. The research is set apart from purely anatomical approaches by two computational aspects. The anatomical knowledge is formalized into abstract mathematical rules tailored for the instantiation of models on supercomputers, and dynamical simulations further constrain the inferred connectivity.
Emphasis is placed on simulations of networks featuring the full biological density of neurons and synapses, in view of inevitable inaccuracies introduced by downscaling. Such full-scale simulations are enabled by the simulation technology of NEST, which runs efficiently on systems including compute clusters and supercomputers. The dependence of the predicted network activity on structural parameters is studied, and the predictions are compared with experimentally recorded activity. With the help of mean-field theory, these comparisons suggest adjustments to the model structure that improve the dynamical predictions. The combination of anatomical analysis, dynamics from spiking simulations, and mean-field theory yields predictions for new anatomical studies and insights into the mechanisms underlying observed brain activity.
large-scale spiking neural network simulations, neuroanatomy, mean-field modeling and theory, primate cerebral cortex