MRI Spectroscopy
The aim of this project is to produce concentration (coloured) maps of brain metabolites. An example can be seen in Fig.2, which shows a map of the concentration ratio between the levels of Creatine (Cr) and N-Acetyl-Aspartate (NAA) in the brain of a tumour-bearing patient.
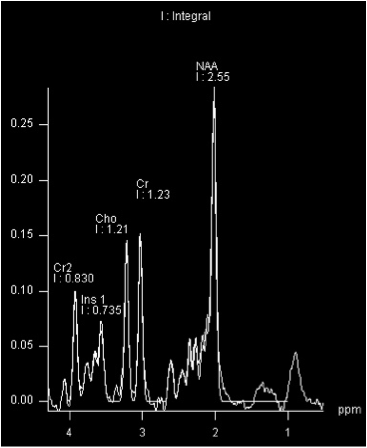
Spectroscopy is a very sensitive MRI technique and a large range of artefacts can corrupt the measured spectrum. However, despite these difficulties, this technique can provide significant, detailed information relating to factors such as the concentrations of relevant chemicals (e.g. neurotransmitters) in the brain. This information can then be used in the diagnosis of brain related diseases and in treatment planning.
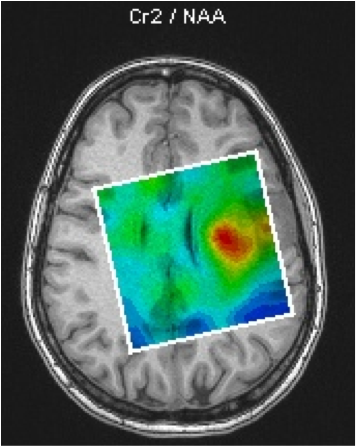
This example of a brain metabolites concentration map was made using the SIEMENS spectroscopy toolbox available on the scanner's console. Such maps can be improved, but in order to be able to produce such maps, a suitable MRI sequence has to be developed. The development of such sequences is a key part of our work.
In order to obtain a coloured concentration map of the brain, the measured spectrum is computed to give the concentration of every metabolite of interest in the spectrum (Fig.1). Several algorithms already exist to do this, and currently, work is being undertaken to improve their accuracy and efficiency. The output of such algorithms is a table stating the computed metabolites concentrations present in the voxels. This information is then transformed into a coloured map.
MR Spectroscopy Imaging (MRSI)
In order to minimise the required scanning time and to improve patient comfort, traditional MR spectroscopy methods acquire MR spectra either from a single voxel or from single slices with straight borders clipped in the brain tissue.
In order to investigate diseases characterised by spatially varying metabolite concentrations, such as brain tumours, it is desirable to measure whole brain spectroscopic data sets with high spatial resolution, while keeping the measurement time low. This is accomplished using the "Echo Planar Spectroscopic Imaging" (EPSI) sequence [Ebel A. and Maudsley A., Magn Reson Med 2005], provided by our co-operation partner Prof. A. Maudsley (University of Miami, FL, USA).
By measuring spatial and spectral information in one acquisition step, the total measurement time can be reduced to 15 min. After the measurement, an automated processing pipeline reconstructs the data, performs spectral fits on all the voxels that show acceptable spectral quality, and yields the corresponding metabolite maps. The various whole brain metabolite distributions can be compared with other imaging modalities, such as 18F-fluoro-ethyl-tyrosine-PET (FET-PET) uptake.
Fig. 3 exemplifies this method for a brain tumour. The low extent of congruency between the spectroscopic tumour markers choline / N-acetyl-acetate (NAA) and FET-uptake requires further analysis and may contribute to prospective, more sensitive and/or specific methods for diagnosis, treatment monitoring and relapse control of brain tumours [Mauler J. et al., J Nucl Med 59, 2017].
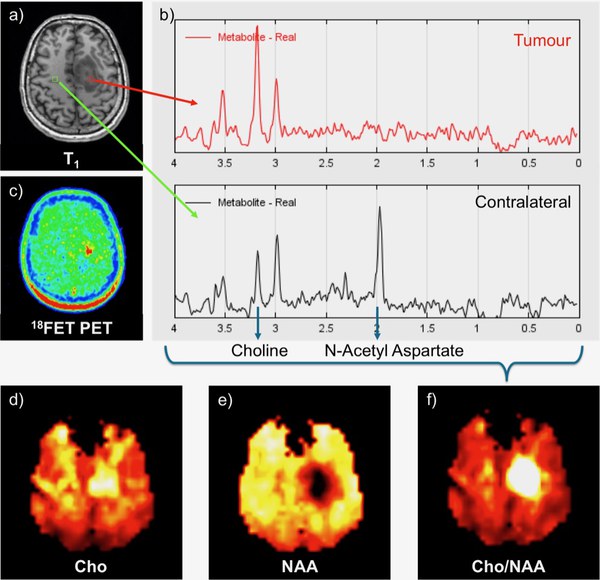
Fig.3 Data acquisition and reconstruction. Astrocytoma in the left gyrus frontalis sup. a) The T1 image shows two exemplarily selected voxels in the tumour and in the contralateral side, which yield the b) corresponding H-MR-spectra. c) The FET-uptake is increased at the position of the lesion. In the tumour tissue, the choline peak is significantly enhanced, whereas the N-acetyl aspartate signal has vanished. The spectra of the full brain data set are transferred into metabolite maps, depicting the distribution of d) Cho and e) NAA. f) The ratio map Cho/NAA clearly shows the lesion [Mauler J. et al., ISMRM 23rd Annual Meeting 2015, Toronto].
