IBI Seminar: Investigating mental illnesses as protein aggregation disorders
Dmitry Fedosov
Nicholas James Bradshaw
Department of Biotechnology, University of Rijeka, Rijeka, Croatia
Join us in person at the PGI Lecture Hall (Building 04.8, Room 365)
Background: Genetic analysis of schizophrenia has made great progress in the last 15 years, identifying many loci that may influence its pathology. However, the existence of many risk variants of small individual effect, combined with rare variants, makes it difficult to identify individual protein targets for downstream study of this devastating psychiatric condition. To complement the genetic approach, we and others have been investigating potential disturbances of proteostasis in schizophrenia. Specifically, we hypothesize that there exist specific proteins that form insoluble protein aggregates in the brains of patients. These would be partially analogous to similar protein accumulations in the neurodegenerative disorders. This symposium will therefore open with a review and update of progress made to date in identifying proteins that aggregate in mental illness.
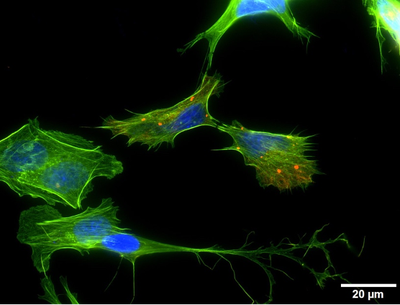
Methods: To date, most candidate aggregating proteins have been determined through the purification of the insoluble protein fraction of postmortem brain tissue (from patients with schizophrenia, bipolar disorder or major depressive disorder, plus controls). These have either been probed for the protein products of classic schizophrenia risk genes, or else subjected to proteomic analysis. Follow up work has largely consisted of expression of these proteins, in wild type, truncated or mutant forms, and examining their expression and effect in cultured cells or primary neurons, through a combination of immunofluorescent microscopy and biochemical techniques.
Results: To date, five proteins have been identified that may form insoluble aggregates in the brains of patients with schizophrenia, and in some instances also in the affective disorders. Of these, DISC1, dysbindin-1 and NPAS3 represent the protein products of classical, pre-GWAS, candidate genes, while CRMP1 and TRIOBP-1 were identified by proteomic approaches, and had not previously been associated with schizophrenia. All are expressed in the brain, and are variously implicated in neurodevelopment and/or synaptic function. Each protein has also been confirmed to be capable of forming aggregates in various experimental systems. We are studying the aggregation of these proteins. Notably, in the case of at least DISC1, TRIOBP-1 and NPAS3, their aggregation propensity is seen to be dependent on specific structural regions of the proteins. Such data allows further analysis of the mechanisms underlying their aggregation, and suggests routes ahead for their study. It is also notable that most of the proteins aggregate independently of each other, with only DISC1 showing a clear propensity to “co-aggregate” with other mental illness proteins in vivo and in vitro.
Conclusions: While still in its infancy as a field, the existence of several insoluble or aggregated proteins in the brains of schizophrenia patients has now been confirmed. Further replication in a wider number of patients is now needed to determine whether this indeed represents a general biological feature of the condition. In parallel, cell and animal studies will help us to understand the pathophysiological consequences of such protein aggregates.
