Bachelor and Master theses
Further opportunities
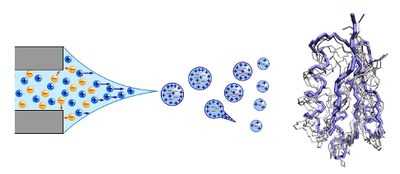
Master thesis on computational mass spectrometry of native proteins
Mass spectrometry (MS) has developed into a central tool of structural biology. In particular, native MS allows to transfer intact molecules in the gas phase, preserving native-like structures and supramolecular complexes. However, the underlying molecular mechanism of protein electrospray in native MS are not yet well understood, hindering structural interpretation of the results. A previous collaboration between the computational group of Prof. Paolo Carloni in Juelich and the experimental group of Prof. Rita Grandori in Milan has implemented a combined protocol by Monte Carlo and molecular dynamics methods to simulate protein structures under electrospray conditions, facing the challenging problem of predicting the most stable protein protomer and providing structural models to interpret the experimental results.
We plan to extend application of this procedure to intrinsically disordered proteins of neurological relevance. As a relevant target to this regard, we will focus on osteocalcin, since it is small and undergoes functionally relevant conformational changes. This protein has pleiotropic functions in the bone, in metabolism and in the brain, which are regulated by carboxylations and calcium binding. Thus, native MS is a promising tool to investigate its conformational ensemble as a function of post-translational modifications and calcium binding.
We are looking for highly motivated individuals with an interest in molecular simulations and biophysics. The research will be held in close collaboration with the experimental lab of Prof. Rita Grandori at the University of Milano-Bicocca.
Contacts: p.carloni@fz-juelich.de and rita.grandori@unimib.it
References
- Ami et al. 2024, J Am Chem Soc 146, 27755-27769. doi: 10.1021/jacs.4c09732
- Li et al. 2017, J Phys Chem Lett 8, 1105-1112. doi: 10.1021/acs.jpclett.7b00127
- Li et al. 2016, Mass Spectrom Rev 35, 111-122. doi: 10.1002/mas.21465
- D’Urzo et al 2015, J Am Soc Mass Spectrom 26, 472-481. doi: 10.1007/s13361-014-1048-z
- Marchese et al. 2012, J Am Soc Mass Spectrom 23, 1903-10. doi: 10.1007/s13361-012-0449-0
- Marchese et al. 2010, PLoS Comput Biol 6, e1000775. doi: 10.1371/journal.pcbi.1000775
