Webservers
GOMoDo and pyGOMoDo
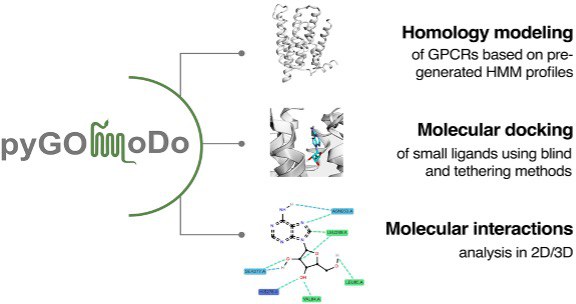
G-protein coupled receptors (GPCRs) are a superfamily of cell signaling membrane proteins that include >750 members in the human genome alone. They are the largest family of drug targets. User-friendly modeling and small molecule docking tools are thus in great demand. While both GPCR structural predictions and docking servers exist separately, with GOMoDo (GPCR Online Modeling and Docking), we provided for several years a web server to seamlessly model GPCR structures and dock ligands to the models in a single consistent pipeline. GOMoDo has been superseded by pyGOMoDo, a python wrap-up of the updated functionalities of the old GOMoDo web server. It was developed having in mind its usage through Jupyter notebooks, where users can create their own protocols of modeling and docking of GPCRs. pyGOMoDo allows performing template choice, homology modeling and either blind or information-driven docking by combining together proven, state of the art bioinformatic tools. The source code is freely available here under the Apache 2.0 license.
References
- M. Sandal et al. GOMoDo: A GPCRs Online Modeling and Docking Webserver. PLoS ONE 8(9), e74092 (2013).
- Rui P. Ribeiro, A. Giorgetti. pyGOMoDo: GPCRs modeling and docking with python. Bioinformatics, (39), Issue 5, btad294 (2023).
CGMD Platform
The Coarse-grained Molecular dynamics(CGMD) platform is a publicly available web server for preparing and running coarse-grained molecular dynamics simulations using different force-fields. The input file is a protein structure. The user is guided through the preparation of the systems, either in a membrane or in solvent, and the running of short simulations following standard protocols.
References
- A. Marchetto et al. CGMD Platform: Integrated Web Servers for the Preparation, Running, and Analysis of Coarse-Grained Molecular Dynamics Simulations. Molecules 2020, 25(24), 5934 (2020).
RepOdor
is conceived with the aim to provide to the users a dynamic and flexible storage tool in the field of olfactory receptors (ORs) and their experimentally known ligands. The goal is to complement the actual landscape of available online resources for Olfaction with a tool that focuses on the interactions between ORs and odorants reported in the literature in the last 30 years.
Stored information about ORs are nomenclature details, organism and sequence, meanwhile information such as physicochemical data, structural features and pharmacological information can be found on odorant molecules. Moreover, information about interaction between ORs and their experimentally known ligands are stored.
Lastly, thanks to the implemented search tools based on BLASTp (for sequence-based receptor similarity) and OpenBabel (for Tanimoto index-based odorant chemical similarity), users will have the chance to find potentially closely related OR-odorant pairs. Thus, RepOdor aims at being a fundamental resource for molecular characterization of OR-odorant pairs, either by reporting already experimentally validated or suggesting new pairs for experimental testing.
MMCG Webserver

The Hybrid Molecular Mechanics/Coarse-Grained (MM/CG) webserver is a server that helps in the preparation and running of multiscale molecular dynamics simulations of G protein-coupled receptors (GPCR) in complex with their ligands.
The Hybrid MM/CG Webserver requires the structure of a GPCR/ligand complex as input and then guides the user through the preparation and running of the simulation.
References
- J. Schneider et al. Hybrid MM/CG Webserver: Automatic Set Up of Molecular Mechanics/Coarse-Grained Simulations for Human G Protein-Coupled Receptor/Ligand Complexes. Front. Mol. Biosci 7, 576689 (2020).

