Protein folding
How do the polypeptide chains which are synthesized at the ribosome find their specific spatial structure? We develop and make use of techniques in order to understand the link between synthesis and the folding of proteins.
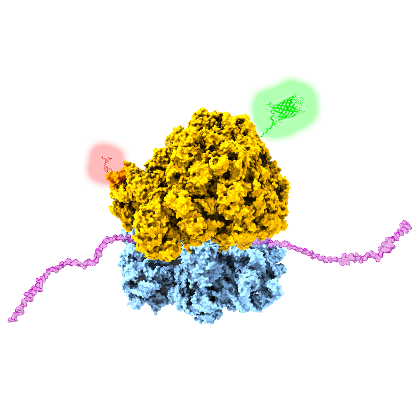
Protein synthesis is a fundamental cellular process, by which ribosomes decode genetic information and convert it into an amino acid sequence. This highly complex process is accomplished by the translational machinery (ribosomes), accessory proteins, tRNA, mRNA and various factors. The fact that protein synthesis and translation does not necessarily require cell integrity, but can also proceed in so called cell-free protein systems, opens the door for comprehensive studies to obtain a deeper understanding of individual steps of the translation cycle and of the folding of de novo synthesized proteins. We made use of this potential by real-time monitoring the green fluorescence protein (GFP) biosynthesis on single molecule level. A suppression of protein release from the ribosome after synthesis and time resolved imaging allowed for the determination of individual GFP maturation times and for quantifying the fraction of active ribosomes in the cell-free reaction format. Furthermore, the overall synthesis performance was quantified with in vitro assays under different environmental conditions (e.g., with/without crowding). In another study we made use of the incorporation of unnatural/modified amino acids into the synthesized polypeptide chain. This approach delivers a powerful method to introduce site-specifically fluorescence labels into proteins, a prerequisite for expanding the scope of single-molecule FRET applications (e.g. co-translational protein folding).
Related Publications
- Cerminara M., Schöne A., Ritter I., Gabba M., Fitter J. (2020) Mapping Multiple Distances in a Multidomain Protein for the Identification of Folding Intermediates Biophys. J, 118, 688-697
- Höfig H., Yukhnovets O., Remes C., Kempf N., Katranidis A., Kempe D., Fitter J. (2019) Brightness-gated two-color coincidence detection unravels two distinct mechanisms in bacterial protein translation initiation. Communications Biology, 2:459
- Sadoine M., Cerminara M., Gerrits M., Fitter J., Katranidis A. (2018) Cotranslational Incorporation into Proteins of a Fluorophore Suitable for smFRETStudies. ACS Synth Biol., 7, 405-411
- Sadoine M., Cerminara M., Kempf N., Gerrits M., Fitter J., A. Katranidis (2017) Selective Double-Labeling of Cell-Free Synthesized Proteins for More Accurate smFRET Studies. Anal. Chem., 89, 11278-11285
- Kempf N., Remes C., Ledesch R., Züchner T., Höfig H., Ritter I., Katranidis A., Fitter J. (2017) A Novel Method to Evaluate Ribosomal Performance in Cell-Free Protein Synthesis Systems. Scientific Reports, 7: 46753
- Lamprou P., Kempe D., Katranidis A., Büldt G., Fitter J. (2014) Nanosecond Dynamics of Calmodulin and Ribosome-Bound Nascent Chains Studied by Time-Resolved Fluorescence Anisotropy. ChemBioChem, 15, 977-985
- Katranidis A., Atta D., Schlesinger R., Nierhaus K.H., Choli-Papadopoulou T., Gregor I., Gerrits M., Büldt G., Fitter J. (2009) Fast biosynthesis of GFP molecules - a single molecule fluorescence study. Angewandte Chemie Int. Edit., 48, 1758-1761
