Arbor
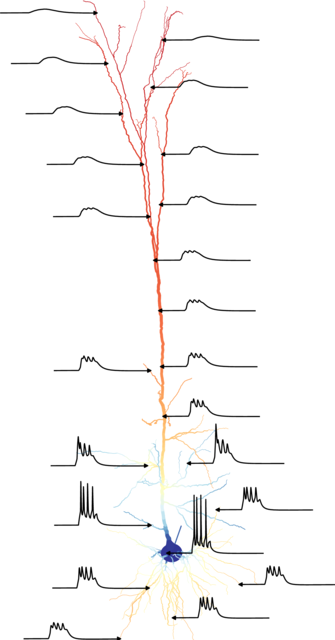
Arbor [1, 2] is a performance-portable library for the simulation of large networks of multi-compartment, morphologically detailed neurons on emerging HPC architectures. It is developed in close collaboration with the neuroscience community in an open model by the Jülich Supercomputing Centre’s Sim- and DataLab Neuroscience and the Swiss National Supercomputing Center (CSCS) and available under a permissive license.
Arbor simulates networks of spiking neurons, particularly multi-compartment neurons, characterized by axonal delays, synaptic and dendritic functions, and the morphological tree. Each cell is modeled as a branching, one-dimensional electrical system with dynamics derived from the balance of transmembrane currents with axial currents that travel through the intracellular medium, and with ion channels and synapses represented by additional current sources.
In networks, interactions between cells is conveyed by spikes and gap junctions. Besides biophysical neural models, Arbor models additional cell kinds, like leaky integrate-and-fire cells and proxy spike sources.
Users work with Arbor via a convenient interface in Python or a lower-level C++ API. Arbor has been shown to excellently scale to large GPU deployments and make good use of hardware capabilities.
The SDLN is involved in the following activities:
- Software development
- Benchmarking and testing
- Scientific studies
- Neuroscience community outreach within the scope of training sessions and workshops
Links
- Source code: https://github.com/arbor-sim/arbor
- Documentation: https://arbor.readthedocs.io
Arbor on HPC
Arbor shows excellent scalability and has been used to assess the performance of various Tier 1 HPC deployments used for neuroscience research.
Background and Motivation
New HPC architectures, such as the addition of ubiquitous GPU resources, have been a new challenge. The evolution of computing equipment ranging from the desktop PCs to supercomputers has enabled a plethora of tools for numerically computing predictions of neuronal network behavior that is comparable with a variety of experimental results, thus allowing the rigorous testing of possible functional models with varying levels of experimental verification, mathematical validity
and stability, and computational performance. New HPC architectures such as the addition of ubiquitous GPU resources have been a new challenge. Developing performant algorithms for computing the Hines matrix on GPUs and other vectorized hardware has been an additional hurdle [3]. The development of Arbor has focused on tackling issues of vectorization and emerging hardware architectures by using modern C++ and automated code generation, within an open-source and open-development model.
Our Approach
Arbor is designed to accommodate these primary goals:
- scalability,
- ease of use,
- extensibility,
- performance portability.
Scalability is achieved through distributed model construction, following the
abstraction of a recipe and through the use of an asynchronous MPI-based spike
communication scheme.
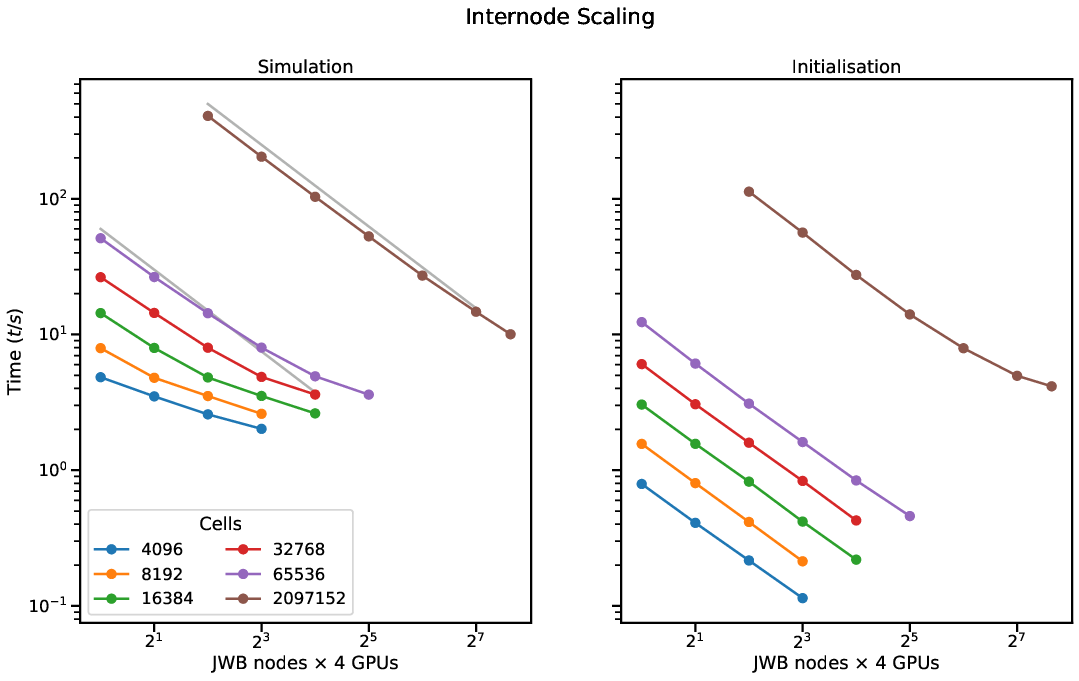
This allows, for example, to leverage CPU and GPU at the same time while still setting aside dedicated CPU resources for off-loading spike processing and communication.To achieve this abstraction, Arbor separates the description of simulations from their execution. First, a ’recipe’ describes a network or cell model This allows for delayed and optimized choices of the distribution of work onto the available hardware. Second, users provide a request for the available computational resources: MPI tasks, threads, and GPUs. Thus, scientific and computational details are separate concerns. Porting a model to a different supported platform requires changing only this — usually small — fraction of the simulation. Finally, A simulation is an executable instantiation of a model, derived from both parts of the description. For each platform — currently multicore CPUs with SIMD (AVX, NEON, SVE), NVIDIA CUDA, and AMD HIP — a specialized and optimized backend is provided. This allows for example to leverage CPU and GPU at the same time while still setting aside dedicated CPU resources for off-loading spike processing and communcation.
Multi-Compartment Neurons
The description of multi-compartment cells include the specification of ion channel and synapse dynamics. A bundled translator is used to compile a subset of NEURON’s specification language NMODL, and layouts can be specified using various well-known file formats (SWC, NeuroLucida ASC, and NML).
Arbor is extensible, allowing for the creation of new kinds of cells and new kinds of cell implementations, while target-specific vectorization, code generation and cell group implementations allow hardware optimized performance of models specified in a portable and generic way.
High-Performance Computing in Arbor
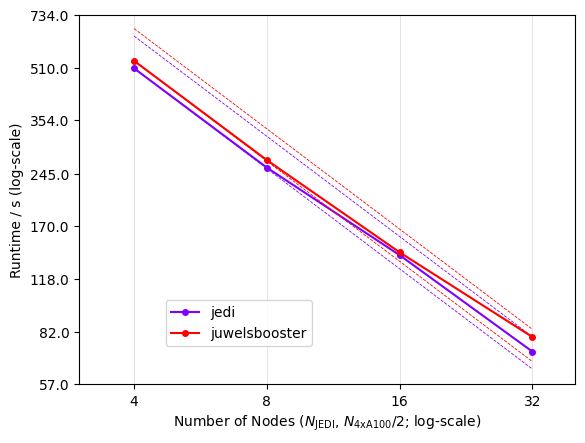
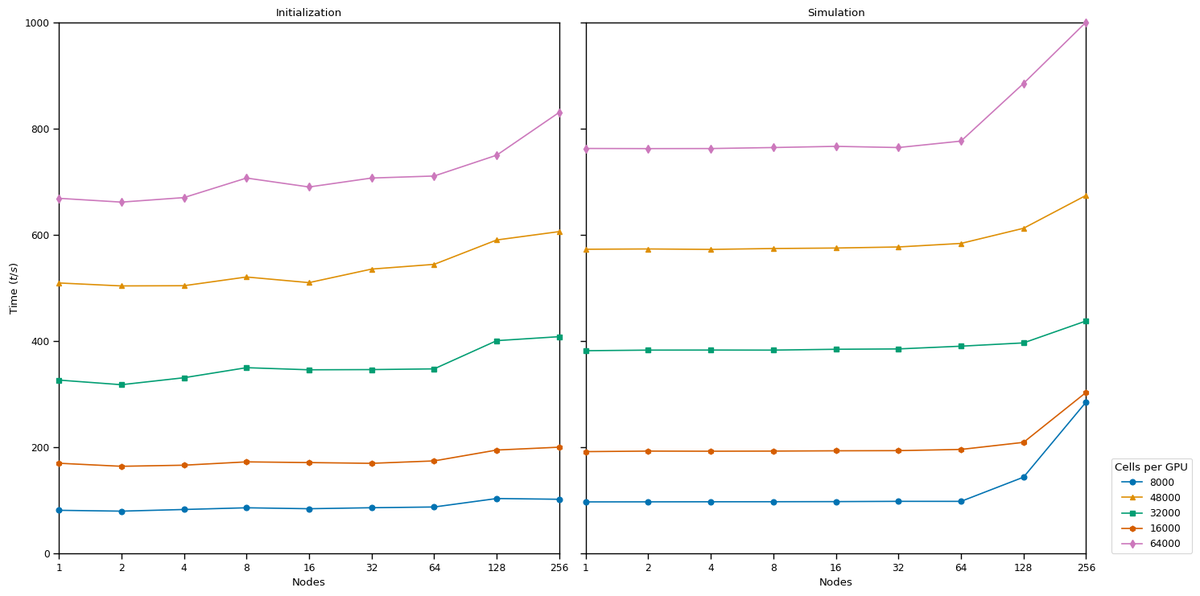
The Arbor library is an active open source project, written in C++17 and CUDA using an open development model. It can scale from laptops to the largest HPC clusters using MPI. The on-node implementation is specialized for GPUs, vectorized multicore, and Intel KNL. Its modular design enables extensibility to new computer architectures, and employs specific optimizations for these GPU and CPU implementations.
Wider Ecosystem
Arbor-GUI is a cross-platform graphical tool for designing and exporting single cell models (https://github.com/arbor-sim/gui).
Benchmarking and validation of Arbor and other simulators can be performed with the NSuite performance and validation suite
(https://github.com/arbor-sim/nsuite).
Support for the SONATA model exchange format is under active development and available as a preview (https://github.com/thorstenhater/cantata).
Similarly, a translator from the NeuroML2 simulation specification to Arbor models is available here (https://github.com/thorstenhater/nmlcc).
Arbor provides APIs for integration with other tools and simulators, including co-simulation with NEST, TVB, and other spiking neural network simulators in general.
Our collaboration partners
Arbor is being developed in collaboration with the Swiss National Supercomputing Center (CSCS).
References
- N. Abi Akar et al., Arbor - A Morphologically-Detailed Neural Network Simulation Library for Contemporary High-Performance Computing Architectures, 2019 27th Euromicro International Conference on Parallel, Distributed and Network-Based Processing (PDP), Pavia, Italy, 13 Feb 2019 - 15 Feb 2019, DOI: 10.1109/EMPDP.2019.8671560
- Cumming, B., Yates, S., Hater, T., Lu, H., Huisman, B., Wouter, K., Bösch, F., Frasch, S., de Schepper, R., & Luboeinski, J. (2024). Arbor v0.10.0 (v0.10.0). Zenodo. https://doi.org/10.5281/zenodo.13284789
- Huber, Felix, Efficient Tree Solver for Hines Matrices on the GPU, arXiv preprint arXiv:1810.12742 (2018).


