Cell Engineering
Cell Engineering Group
About
By introducing and specifically targeting new proteins into cells, we aim to create systems that allow us to probe information processing in cell networks and generate new bioelectronic materials.
We compare the spatial specificity and persistence of information when a neural network is manipulated using different modalities. Mechanical stimulation, electrical stimulation, and optogenetic stimulation are all candidates for future corrective therapies modulating neuronal activity. Our work underpins the predictions that will be necessary to select the most efficient means of stimulation for a given purpose and to code information correctly.
Research Topics
Our current research is focused on:
- Optogenetic Actuators
- Specificity of stimulation
- Identifying and Manipulating Key Neurons.
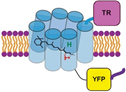
Stimulation and inhibition of neuronal signaling using light actuated channels and pumps. We utilize optogenetic actuators based on Channelrhodopsin and Anionchannelrhodopsin to manipulate neuronal networks at the single cell level with high flexibility in both spatial and temporal protocols. To expand the usefulness of optogenetic actuators, we explore tagging domains capable of subcellularly targeting the actuators and allowing their use in other cell types, such as epithelia (Fig. 1).
Genetically Encoded Calcium Indicators (GECIs)
The membrane potential of the cell is monitored fluorescently using GcAMP or RcAMP. Though not as temporally accurate as electrical methods, GECIs provide signaling information over sizable areas without gaps in recording areas, or difficult-to-attribute multiple unit detection on a single pixel.
Specificity of Stimulation
Even when targeting a single neuron, the method of stimulation can cause a network response of varying extent and duration. For example, a single mechanical stimulation of a single neuron can affect signaling in cells hundreds of micrometers away and alter network activity for minutes. Extracellular electrical stimulation is fast, but faces challenges of neurites crossing through the generated field. We are comparing these methods with optogenetic single cell stimulation to optimize stimulation modes for desired activity patterns.
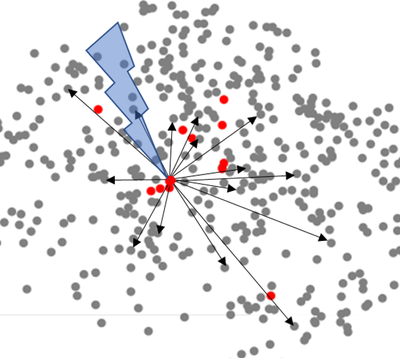
Not all neurons in a network have equal ability to influence signaling in the network. We are working to identify these key influencers and how different modes of manipulation at the single cell level can alter wider network signaling. This will provide the most efficient methods for correcting aberrant network activity (as in disease or injury) or for programming information into the network (as in biomimetic computing) (Fig. 2).
Members
Former Members
Devran Dardanoglu, intern until 08/2023
Bogdana Cepkenovic, Doctoral researcher until 04/2023
Ruoyan Wei , Doctoral researcher until 12/2022
Jiali Wang , Doctoral researcher until 12/2022
Cole Wilson, Fulbright Fellow until 07/2022
Dominik Brinkmann, Doctoral researcher until 12/2021
Timm Hondrich, Doctoral Researcher until 09/2020
Lucas Bertram, Master Student until 12/2019
Jana Schieren, Master Student until 03/2019
Irina Tihaa, Doctoral Researcher until 06/2018
Annika Graeve, Bachelor Student until 03/2018
Sarah Roßbiegalle, Bachelor Student until 02/2018
Wenfang Li, Doctoral researcher until 03/2017
Lei Jin, Doctoral Researcher until 07/2016
Recent Publications
- Expressing Optogenetic Actuators Fused to N‐terminal Mucin Motifs Delivers Targets to Specific Subcellular Compartments in Polarized Cells. Wang, J., Platz‐Baudin, E., Noetzel, E., Offenhäusser, A., & Maybeck, V. (2024). Advanced Biology, 8(3).
- Single-neuron mechanical perturbation evokes calcium plateaus that excite and modulate the network. Cepkenovic, B., Friedland, F., Noetzel, E., Maybeck, V., & Offenhäusser, A. (2023). Scientific Reports, 13(1), 20669. doi.org/10.1038/s41598-023-47090-z
- Kempmann, A., Gensch, T., Offenhäusser, A., Tihaa, I., Maybeck, V., Balfanz, S., & Baumann, A. (2022). The Functional Characterization of GCaMP3.0 Variants Specifically Targeted to Subcellular Domains. International Journal of Molecular Sciences, 23(12). https://doi.org/10.3390/ijms23126593
- Shokoohimehr, P., Cepkenovic, B., Milos, F., Bednár, J., Hassani, H., Maybeck, V., & Offenhäusser, A. (2022). High‐Aspect‐Ratio Nanoelectrodes Enable Long‐Term Recordings of Neuronal Signals with Subthreshold Resolution. Small, 2200053. https://doi.org/10.1002/smll.202200053
- Improvements of Microcontact Printing for Micropatterned Cell Growth by Contrast Enhancement, Hondrich et al., Micromachines 2019, 10, 659; doi:10.3390/mi10100659, https://www.mdpi.com/2072-666X/10/10/659
- How to image cell adhesion on soft polymers? Seyock et al., Micron 2017, http://dx.doi.org/10.1016/j.micron.2016.11.002
- Controlled Engineering of Oxide Surfaces for Bioelectronics Applications Using Organic Mixed Monolayers, Markov et al., ACS Applied Materials and Interfaces 2017 10.1021/acsami.7b08481, https://pubs.acs.org/doi/abs/10.1021/acsami.7b08481
- High-efficiency transduction and specific expression of ChR2opt for optogenetic manipulation of primary cortical neurons mediated by recombinant adeno-associated viruses, Jin et al., J Biotechnol,2016, 233:171–80, https://doi.org/10.1016/j.jbiotec.2016.07.001
- An evaluation of extracellular MEA versus optogenetic stimulation of cortical neurons. Maybeck et al., Biomed Phys Eng Express, 2016, 2:055017, https://doi.org/10.1088/2057-1976/2/5/055017