Carsten Sachse
Forschungszentrum Jülich

Structural biology of essential cellular processes using electron cryo-microscopy (cryo-EM); biochemically purified macromolecular structures and multi-protein complexes being investigated in their near-native environment in vitro or inside cells without the need for crystallization; autophagy; software development for protein structure determination.
Vita
since 2018 | Professor of Biology at Heinrich Heine University Düsseldorf |
|---|---|
since 2017 | Director of the Institute for Structural Biology at the Ernst Ruska-Centre for Microscopy and Spectroscopy with Electrons, Forschungszentrum Jülich |
2010-2018 | Group leader at the Structural and Computational Biology Unit of the European Molecular Biology Laboratory, Heidelberg |
2008-2009 | Postdoctoral fellow with Roger Williams and Richard Henderson at the MRC Laboratory of Molecular Biology, Cambridge (UK) |
2007-2008 | Postdoctoral scientist with Marcus Fändrich at the Max-Planck-Research Unit “Enzymology of Protein Folding”, Halle |
2007 | Ph. D. at Friedrich Schiller University Jena |
2004-2007 | Ph. D. project on high-resolution electron cryomicroscopy of amyloid fibrils in the groups of Marcus Fändrich (Leibniz Institute for Age Research, Jena; now in Ulm) and Niko Grigorieff (Brandeis University, U.S.A.; now at Janelia Farm, U.S.A.) |
2001-2003 | Study of Biochemistry at Friedrich Schiller University Jena |
Research
Autophagy (from the Greek, meaning ‘to eat oneself’) is the cell’s housekeeping mechanism to engulf and degrade long-lived proteins, macromolecular aggregates, damaged organelles and even microbes in double-membrane vesicles called autophagosomes.
In our group, we investigate the molecular structures involved in autophagy as they provide fundamental insights for our understanding of aberrant cellular processes like cancer, ageing or infection.

Multiprotein complexes are essential mediators in the events leading to autophagy. On the structural level, however, little is known about their 3D architecture. Fundamental questions on the nature of these complexes need to be addressed:
- How are protein deposits structurally linked to autophagy?
- What are the shapes of these multiprotein assemblies at the membrane?
- How do they give rise to the cellular structure of the autophagosome?
Electron cryomicroscopy is a powerful structural biology technique
We study the structures of molecular assemblies using biochemical and biophysical techniques, and subsequently visualise them by electron cryomicroscopy (cryo-EM). By this technique, large macromolecular structures and multi-protein complexes can be studied in their near-native environment without the need for crystalisation. Small amounts of material are sufficient to obtain ‘snapshots’ of single particles in the electron cryomicroscope. The molecular images are combined by computer-aided image processing techniques to compute their 3D structures. As recent advances in hardware and software have led to a wave of atomic-resolution structures, cryo-EM shows great promise in becoming a routine tool for high-resolution structure determination of large macromolecules. To further realise the potential of the technique, the scientific community is still in great need of hardware-based improvements and software enhancements. Therefore, we are also interested in developing techniques, including sample preparation and data processing, to routinely achieve atomic-resolution structures by single-particle cryo-EM. For example, in our group we actively develop the software SPRING for high-resolution cryo-EM structure determination of specimens with helical symmetry.
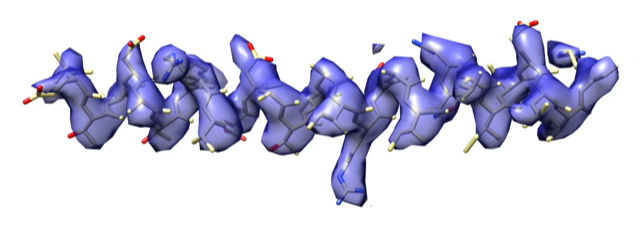
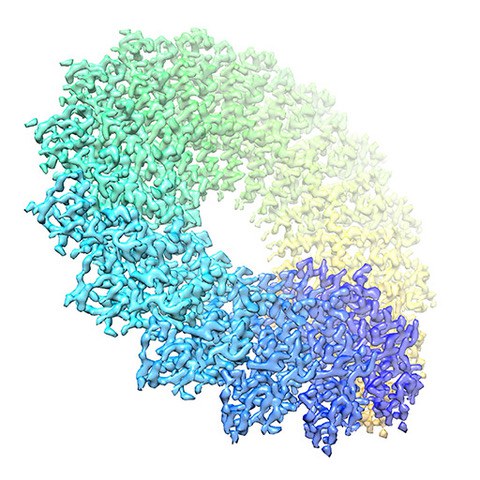
Method development
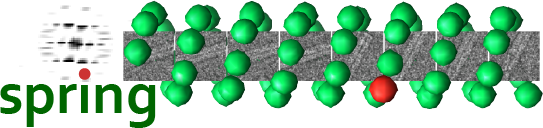
- Spring
- Helical diffraction simulator
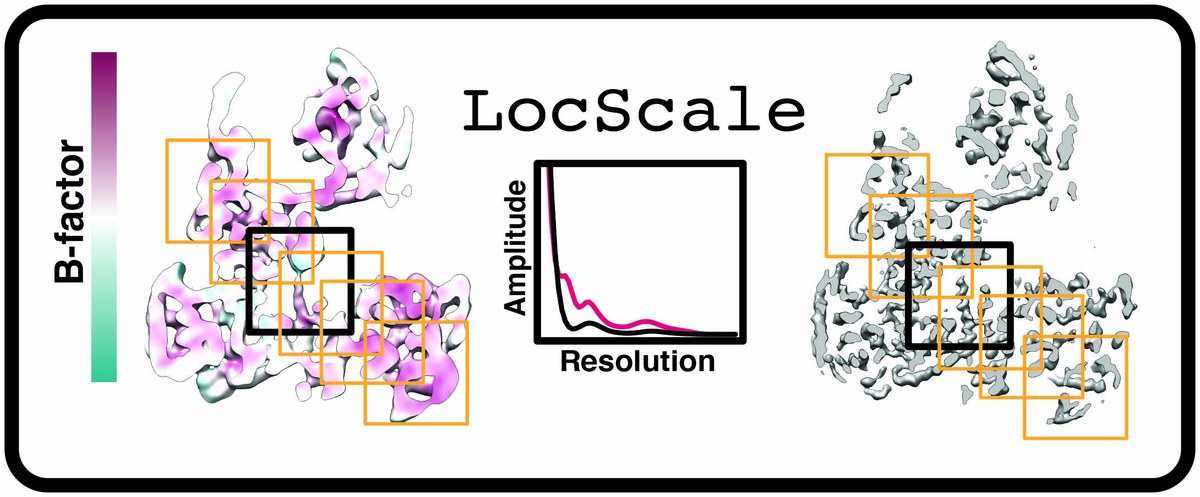
- LocScale
Spring is a single-particle based helical reconstruction package developed in our laboratory to determine 3D structures of a variety of highly ordered and less ordered specimens with helical symmetry from electron micrographs.
Helical diffraction simulator is a web server tool developed in our laboratory to display Fourier patterns of helices based on input helical symmetry parameters according to the theory formulated by Cochran, Crick and Vand in 1952.

LocScale is density scaling (sharpening) procedure that aims to enhance interpretability of cryo-EM density maps. LocScale is a reference-based local amplitude scaling tool using prior model information to improve contrast of cryo-EM density maps.