Example from Gluconobacter oxydans: Expression systems
The α-proteobacterium G. oxydans is a Gram-negative acetic acid bacterium used for a broad range of industrial applications due to its ability to oxidize a great variety of carbohydrates in the periplasm. In biotechnological applications and basic research, timed production of proteins in bacterial cell culture for various purposes by inducible or regulatable expression of target genes is a standard method well established in many bacteria except in acetic acid bacteria.
See also our review:
Fricke P.M., Klemm A., Bott M., Polen T. (2021)
On the way toward regulatable expression systems in acetic acid bacteria: target gene expression and use cases.
Appl Microbiol Biotechnol 105(9):3423-3456
doi: 10.1007/s00253-021-11269-z
Based on our previous findings on regulatable expressions systems (see below) we are now on the way to analyse and establish an optogenetic toolbox for light-controlled gene expression in Gluconobacter in a running BioSC project:
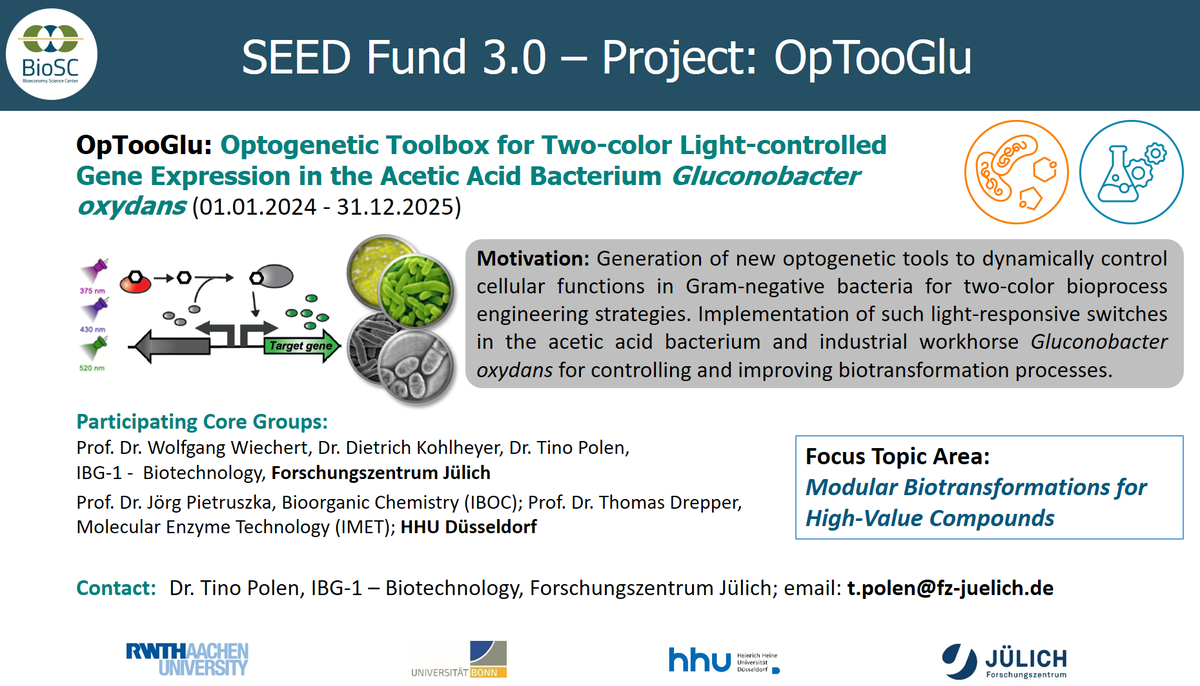
Recently, we found that the L-arabinose-inducible ParaBAD promoter and the transcriptional regulator AraC from E. coli MC4100 perform very well in G. oxydans with a pBBR1MCS-5 backbone allowing up to 480-fold induction with good tunability from 0.1 to 1% L-arabinose, demonstrating superior functionality of an AraC-ParaBAD system in G. oxydans. This represents the first system for regulatable target gene expression in G. oxydans, which could potentially be used in other AAB.
Fricke P.M., Link T., Gätgens J., Sonntag C., Otto M., Bott M., Polen T. (2020)
A tunable l-arabinose-inducible expression plasmid for the acetic acid bacterium Gluconobacter oxydans.
Appl Microbiol Biotechnol 104(21):9267-9282
doi:10.1007/s00253-020-10905-4
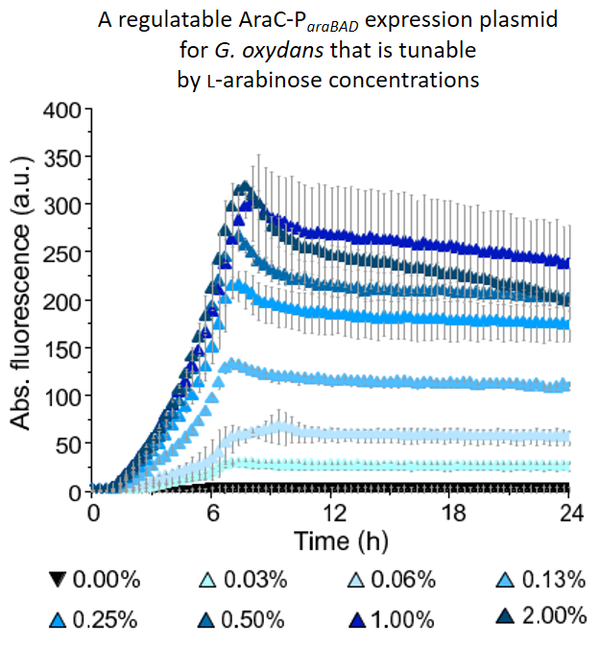
Furthermore, we found with a pBBR1MCS-5-based TetR-Ptet system that inducible target gene expression based solely on de-repression of the heterologous target promoter can also perform extremely well in G. oxydans. With the pBBR1MCS-5-based plasmid, the anhydrotetracycline (ATc)-inducible promoter Ptet derived from the E. coli transposon Tn10 exhibited excellently tunable expression performance in an inducer concentration-dependent manner with maximal induction ratios up to more than 3,500-fold. This was due to extremely low basal reporter expression in the absence of an inducer, thus well repressed Ptet, and Ptet being very strong in G. oxydans.
Plasmid-based mNeonGreen fluorescence reporter protein expression was gradually inducible by increasing ATc concentrations and the induction was highly homogeneous according to FACS results:
Fricke P.M., Lürkens M., Hünnefeld M., Sonntag C.K., Bott M., Davari M.D., Polen T. (2021)
Highly tunable TetR-dependent target gene expression in the acetic acid bacterium Gluconobacter oxydans.
Appl Microbiol Biotechnol 105(18):6835-6852
doi:10.1007/s00253-021-11473-x
In G. oxydans, the high periplasmic oxidation of substrates combined with a low carbon flux into the cytoplasmic metabolism having no genes encoding for phosphofructokinase, succinyl-CoA synthetase, and succinate dehydrogenase typically results in a low biomass yield which limits a broader range of industrial applications. Recently, heterologous genes were chromosomally integrated and expressed to complete the TCA cycle resulting in about 60% increased biomass yield (PMID 28484812).
Enforcing the cytoplasmic glucose catabolism in G. oxydans in such a way may cause suppressor mutations and structural variants affecting the genome stability. To check this we conducted a hybrid approach for genome sequencing by combining Oxford Nanopore’s MinION® sequencing for long reads and Illumina’s technology for short accurate reads. The genome sequence of the engineered G. oxydans strain was stable at least up to 70 generations of strain handling including process time in a controlled bioreactor. The long read data also revealed a novel 1420 bp transposon-flanked and ORF-containing sequence which was hitherto unknown in the G. oxydans 621H reference. Further analysis and genome sequencing showed that this region is also already present in G. oxydans 621H wild-type strains.
Sequencing of primary transcriptomes of G. oxydans 621H revealed 2449 TSSs, which were classified according to their genomic context followed by identification of promoter and ribosome binding site motifs, analysis of 5´-UTRs including validation of predicted cis-regulatory elements and correction of start codons. 1144 (41%) of all genes were found to be expressed monocistronically, whereas 1634 genes were organized in 571 operons. Together, TSSs and whole transcriptome data revealed 18 novel intergenic, 328 intragenic, and 313 antisense transcripts.
The estimated mRNA half-lives in G. oxydans overall ranged mainly from 3 min to 25 min with a global mean of 5.7 min. The transcripts encoding GroES and GroEL required for proper protein folding ranked at the top among transcripts exhibiting both long half-lives and high abundance. The F-type H+-ATP synthase transcripts involved in energy metabolism ranked among the transcripts with the shortest mRNA half-lives. Also, RNAseq analysis revealed low expression levels for genes of the incomplete TCA cycle and also the mRNA half-lives of several of those were short and below the global mean.
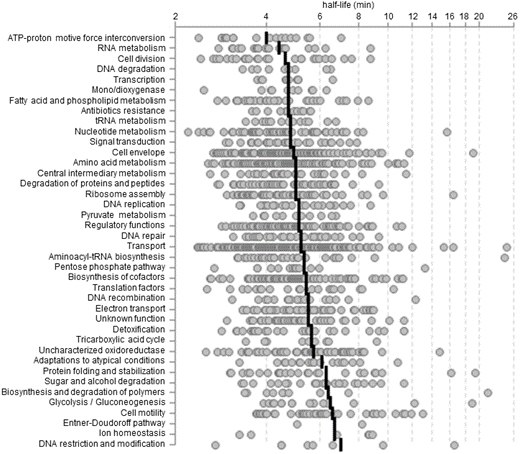
The mRNA decay analysis also revealed an apparent instability of full-length 23S rRNA. Further analysis of the ribosome-associated rRNA revealed a 23S rRNA fragmentation pattern exhibiting new cleavage regions in 23S rRNAs which were previously not known.
Kranz A., Vogel A., Degner U., Kiefler I., Bott M., Usadel B., Polen T. (2017)
High precision genome sequencing of engineered Gluconobacter oxydans 621H by combining long nanopore and short accurate Illumina reads.
J Biotechnol pii: S0168-1656(17)30170-0
doi: 10.1016/j.jbiotec.2017.04.016
Kranz A., Busche T., Vogel A., Usadel B., Kalinowski J., Bott M., Polen T. (2018)
RNAseq analysis of alpha-proteobacterium Gluconobacter oxydans 621H.
BMC Genomics 19(1):24
doi: 10.1186/s12864-017-4415-x
Kranz A., Steinmann A., Degner U., Mengus-Kaya A., Matamouros S., Bott M.,
Polen T. (2018)
Global mRNA decay and 23S rRNA fragmentation in Gluconobacter oxydans 621H.
BMC Genomics 19(1):753
doi: 10.1186/s12864-018-5111-1
