Research
Corynebacterium glutamicum - systemic understanding of an industrial workhorse
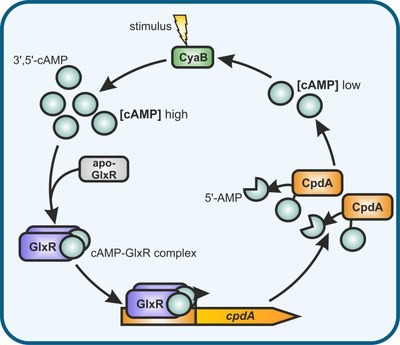
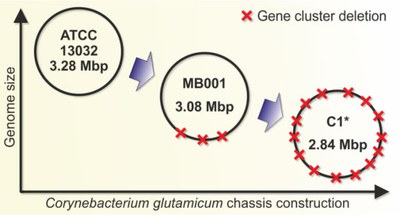
Due to their unsurpassed capabilities in catalysing multistep biosynthetic pathways rapidly and efficiently, microbial cell factories (MCFs) are our major systems for transformation of renewable carbon sources into value-added products. For rational strain development, systemic and detailed knowledge of the metabolic and regulatory network is an essential prerequisite, and fundamental studies on these topics are an important part of our research. Our group focuses on Corynebacterium glutamicum, an industrial amino acid and protein producer with superior robustness and a strong perspective to become a major production platform for a variety of low and high-value products such as bio-monomers and pharmaceuticals. We are especially interested in the understanding of regulatory networks on different levels and the investigation of essential genes of so far unknown function. In this context we also created the C. glutamicum chassis strain C1* with a 13.4% reduced genome which still shows wild type-like behaviour under defined conditions. More Information
Transcription factor-based biosensors - accelerating metabolic and enzyme engineering

Genetically encoded single-cell biosensors provide solutions to some very important challenges in bioprocess development as well as some fundamental questions in microbiology. We use our knowledge about natural regulatory processes to construct genetically encoded biosensors that are able to detect e.g. an intracellular metabolite concentration and convert this information into an easily measurable fluorescence output. These biosensors allow high-throughput screening of genome-wide, gene-specific or site-specific mutant libraries using fluorescence activated cell sorting (FACS) to isolate single cells with e.g. increased fluorescence, representing strain or enzyme candidates with beneficial mutations. These clones are then further characterized for example by microcultivation systems (Biolector), HPLC analysis, and genome analysis. Our in-house sequencing facilities allow a fast identification of potentially beneficial mutations in whole strains. Unfortunately, suitable biosensors are not available for all compounds of interest. Therefore we aim to alter the specificity of biosensors to broaden the spectrum of potential ligands. More Information
Microbial cell factories - efficient production of value added compounds
A major aim of sustainable bioeconomy is a gradual change from fossil to renewable carbon sources in the chemical industry. Cells and enzymes have been optimized for the metabolism of natural carbon sources for more than three billion years. This explains the power of biological catalysts for the conversion of renewable carbon sources to chemicals. Microorganisms are unrivalled particularly when the synthesis path from a given substrate to a desired product requires a large number of reaction steps. Metabolites such as L-glutamate or L-lysine are produced with microbes in the million-ton scale, clearly demonstrating the industrial applicability of bioprocesses also for bulk products. We apply the knowledge gained by our fundamental research and novel tools such as the genetically encoded biosensors to improve microbial strain development using Corynebacterium glutamicum and Gluconobacter oxydans as cell factories. More Information
Methods and Equipment
Our institute is equipped with state of the art technical instruments allowing the application of a wide range of methods:
- Molecular microbiology: standard cloning techniques, site-directed mutagenesis, Gibson assembly
- Protein biochemistry: ÄktaPure, Tecan Infinite M1000Pro, MALDI, Ultracentrifuge
- Interaction analysis: EMSAs, Biacore, ITC, Nanotemper,
- Cultivation at different scale: Biolector, Shaker with humidity control, DasBox
- OMICS techniques: in particular transcriptomics (microarrays & RNA-Seq) and proteomics (NanoLC-MSMS)
- Analytics: uHPLC, LC-MS
- Next-generation sequencing: Genome re-sequencing, ChapSeq (Illumina, MiSeq)
Joining us
We frequently offer Master and PhD positions. Please check the IBG-1 website for open positions or send us your unsolicited application including CV, motivation letter and certificates via email to m.bott@fz-juelich.de.
