Development of Transcription Factor-based Biosensors and their Utilization for High-throughput Screening of Microorganisms using FACS
Metabolic engineering for the microbial overproduction of high-value small molecules is a highly complex process, which is dictated by a large number of parameters. The biosynthetic pathways leading to these products comprise multiple native or heterologous catalytic steps, each one representing a potential bottleneck when directing carbon flux toward target small-molecule production. Furthermore, the host organism’s native genetic regulation network and the interaction with the target pathway can impact final product yields.
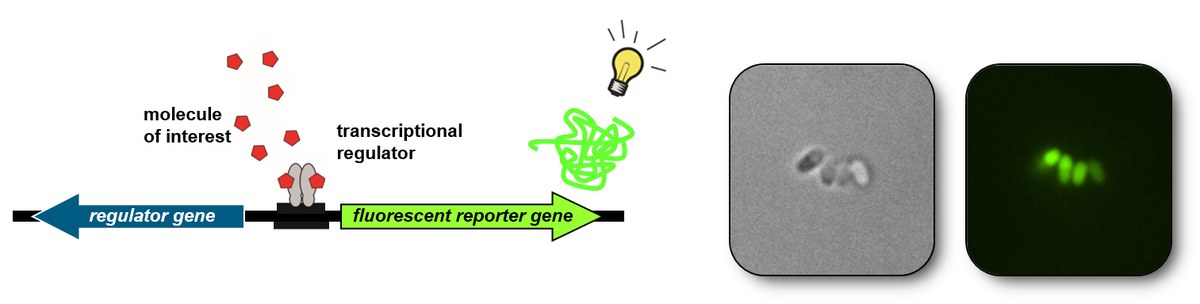
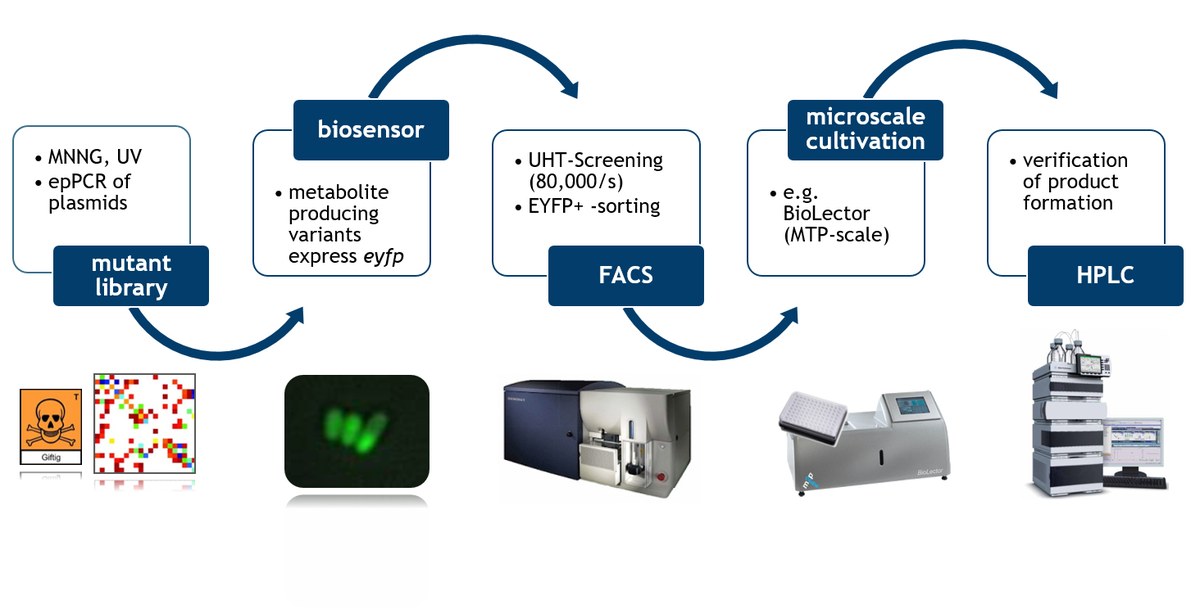
Efforts in the field of metabolic engineering are currently defined by the implementation of a series of rational design-based strategies to modify the host genome and pathway enzymes to achieve moderate product titers. To generate microbial production strains with higher productivities, we want to follow the long trail of successes in protein engineering and develop approaches to high-throughput screening of single cells. For this purpose, we design, construct and apply transcriptional regulators sensing molecules of biotechnological interest, which, in response to this signal, drive transcription of a fluorescent protein [1]. Furthermore, we adapt important biosensor parameters such as specificity, sensitivity and transferability to other microbial hosts [2 - 4].
References:
[1] Eggeling L., Bott M., Marienhagen J. (2015). Novel screening methods - biosensors. Curr. Opin. Biotechnol. 35: 30-36. (http://dx.doi.org/10.1016/j.copbio.2014.12.021)
[2] Flachbart L. K., Sokolowsky S., Marienhagen J. (2019). Displaced by deceivers - Prevention of biosensor cross-talk is pivotal for successful biosensor-based high-throughput screening campaigns. ACS Syn. Biol. 8 (8): 1847-1857. (https://doi.org/10.1021/acssynbio.9b00149)
[3] Della Corte D., van Beek H.L., Syberg F., Schallmey M., Tobola F., Cormann K.U., Schlicker C., Baumann P.T., Krumbach K., Sokolowsky S., Morris C.J., Grünberger A., Hofmann E., Schröder G.F., Marienhagen J. (2020). Engineering and application of a biosensor with focused ligand specificity. Nat. Commun. 11: 4851. (https://doi.org/10.1038/s41467-020-18400-0)
[4] Flachbart L.K., Gertzen C.G.W., Gohlke H., Marienhagen J. (2021). Development of a biosensor platform for phenolic compounds using a transition ligand strategy. ACS Syn. Biol. 10: 2002-2014. (https://doi.org/10.1021/acssynbio.1c00165)
[5] Baumann P.T., Dal Molin M., Aring H., Krumbach K., Müller M.-F., Vroling B., van Summeren-Wesenhagen P.V., Noack N., Marienhagen J. (2023). Beyond rational - biosensor-guided isolation of 100 independently evolved bacterial strain variants and comparative analysis of their genomes. BMC Biology 21:183. (https://doi.org/10.1186/s12915-023-01688-x)
