MibiNet – Race for Iron: Impact of siderophore-networking on spatial interactions in defined communities
Iron is one of the four most abundant elements on earth and it was even claimed to be involved in the origin of life. This trace element is a physiological requirement of every living organism, playing important roles in numerous biological processes. However, due to the extreme low solubility of Fe3+ in aerobic environments and the potential toxicity through the generation of oxidative stress, microbes need to employ sophisticated economic strategies to manage homeostasis of this essential element. This compromises complex logistics such as production, secretion and uptake of Fe3+-complexing siderophores.
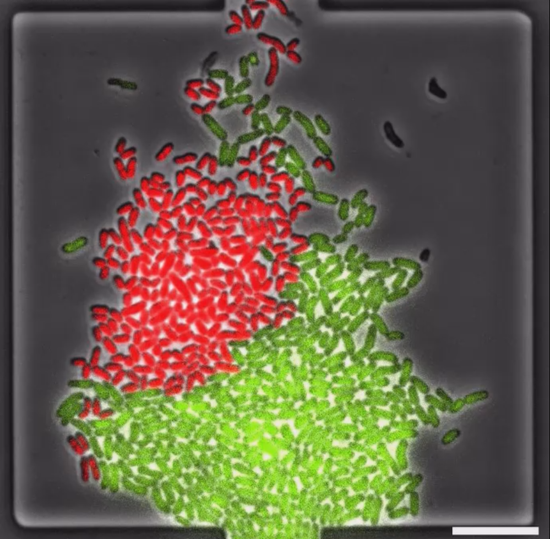
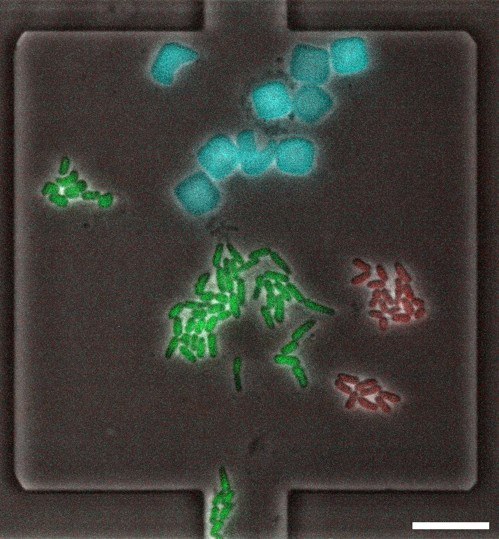
Within this project, we assess competitive and cooperative strategies for the acquisition of iron via siderophores in synthetic microbial communities. These consortia encompass Escherichia coli and Pseudomonas putida, which produce the siderophores enterobactin and pyoverdine, respectively, as well as further microbial strains that specialize in utilizing xeno-siderophores. In order to mimic natural physically and chemically structured microenvironments as well as dynamic changes in the physiology of producers and utilizers, we make use of microfluidic cultivation devices and optogenetic tools. Our goal is to examine the impact of iron management strategies on the structure and dynamics of microbial communities, uncovering both spatial and temporal variations.
Microgels – A Game Changer in Microfluidic Single-Cell Analysis
Spatial bursts in nutrient availability, such as carbon or iron sources, are key drivers of spatiotemporal dynamics in microbial communities. Consequently, the position of individual cells and the resulting neighborhood effects ultimately have a significant impact on cellular growth outcomes. To date, these effects can be studied in a static environment using microfluidic single cell analysis. However, the natural environment of cells is dynamic. Therefore, complex microfluidic environments are required to mimic natural cell behavior. Dynamic control of environmental conditions, such as nutrient concentrations, allows more accurate replication and observation of physiological conditions. Structured environments within a microfluidic chip are therefore urgently needed to understand cellular behavior and interactions, such as iron homeostasis. Therefore, we developed a new approach allowing the integration of microgels inside of the microfluidic growth chambers, whose tunable structure can be used to incorporate and release nutrients and minerals such as iron and glucose in respond to different stimuli (e.g. microgel degrading enzymes; degradation of a dextran-based gel by dextranase), providing a versatile platform for structured microfluidic environments.