RCNMA/NMSim
RCNMA/NMSim is a normal mode-based geometric simulation approach for exploring biologically relevant conformational transitions in proteins. This method is based on a three-step approach for multi-scale modeling of macromolecular conformational changes. The first two steps are based on a rigid cluster normal-mode analysis (RCNMA). In the final step (NMSim), new macromolecule conformers are generated by deforming the structure along low-energy normal mode directions predicted by RCNMA.
A webserver is available here.
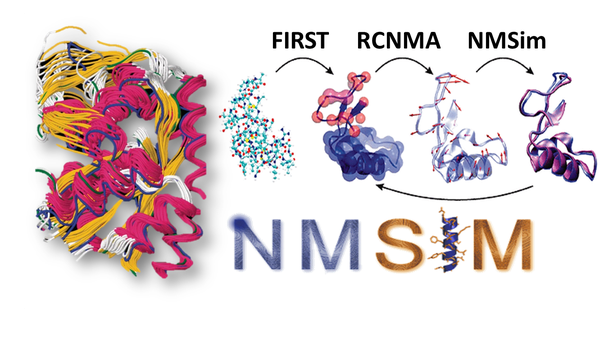
Ahmed et al. J. Chem. Inf. Model. (2011)
Dimura et al. Curr Opin Struct Biol (2016)
Ciupka & Gohlke Scientific Reports (2017)
Last Modified: 01.06.2022
