How Cells Stay Clean and Tidy
Jülich, 23.01.2020. Tidying up and throwing away unwanted items is currently very much in vogue and can even, according to the author of a current bestseller, change lives. Without a doubt, regular clear-outs are also vital for cells: proteins and cell organelles that have become too old and no longer function properly must make way for new ones in due course. If this process is disturbed, cells may be damaged and neurodegenerative diseases can develop. A team of researchers from Germany and Norway has now published new findings on the cellular cleaning mechanism.
Cells dispose of their waste in a two-stage process called autophagy. Cell debris is first collected in bubble-like vesicles known as autophagosomes. Next, these autophagosomes are delivered over to lysosomes, membrane-bound sacs in which specialized enzymes break down the cell waste.
Although the broad outline of this process is well known, the precise details of how this happens have so far remained unclear. New research by scientists from Jülich, Heidelberg, Hamburg and Norway has focused on the role of a protein called p62, which plays an important role in autophagy by delivering cell waste to autophagosomes.
Previous studies have shown that p62 consists of three domains, each with a different role. The PB1 domain enables individual p62 proteins to link together to form long chains. Another, the UBA domain, recognizes special protein tags called polyubiquitin that adhere to proteins and organelles that need to be removed. The third domain, LIR, enables p62 to bind to autophagosomes, ensuring that any cargo bound to UBA ends up in the autophagosome.
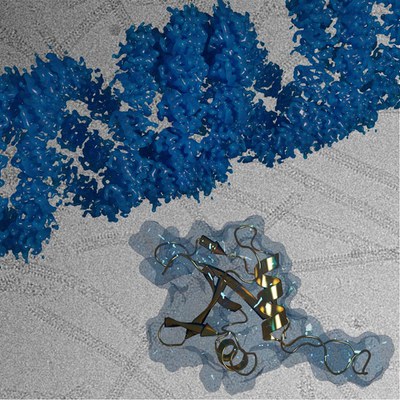
Prof. Carsten Sachse, Director at the Ernst Ruska Centre for Microscopy and Spectroscopy with Electrons in Jülich, together with his colleagues, has now shown that the polymerization of p62 proteins into chains is essential for protein degradation via autophagy. They showed that even smaller polymer chains are sufficient to bring autophagosomes and lysosomes together, but not individual proteins. Additionally, the researchers used cryo-electron microscopy to image the winding, scaffold-like structure of the chains with atomic resolution for the first time. The technique is well suited to shedding light on the structure of large proteins without potentially altering them through the use of preparation measures necessary when using other methods. Studies undertaken on different p62 subtypes of thale cress (Arabidopsis thaliana), also known as mouse-ear cress, and on human cells suggest that the process in plant and animal cells is the same.
Original Publication:
Structural basis of p62/SQSTM1 helical filaments and their role in cellular cargo uptake.
Nat Commun 11, 440 (2020). DOI:
10.1038/s41467-020-14343-8
Further Information:
Website of the Ernst Ruska-Centre for Microscopy and Spectroscopy with Electrons – Structural Biology (ER-C-3)
Website ot the Sachse Lab
Kontakt:
Prof. Dr. Carsten Sachse
Ernst Ruska-Centrum für Mikroskopie und Spektroskopie mit Elektronen – Strukturbiologie (ER-C-3)
Tel: +49 2461 61-2030
E-Mail:
c.sachse@fz-juelich.de
Pressekontakt:
Angela Wenzik, Wissenschaftsjournalistin
Forschungszentrum Jülich
Tel: +49 2461 61-6048
E-Mail:
a.wenzik@fz-juelich.de
